CyEVEX Plugin
CyEVEX is a Cytoscape plugin designed for retrieving gene interactions from EVEX. The plugin enables searching of pairwise interactions for a set of genes or expanding a network with the interaction partners of a given gene.
CyEVEX is distributed as a Java archive which can be downloaded from here: CyEVEX_v0.1.1.jar.
Instructions
Installation
CyEVEX plugin has been tested on Cytoscape version 2.8.3. Download the CyEVEX .jar file and place it into the plugins folder of your Cytoscape installation. Restart Cytoscape. A new submenu called CyEVEX should appear under the Plugins menu.
Usage
Searching edges for existing nodes. Select the nodes you want to include in the search and choose Plugins -> CyEVEX -> Search edges from EVEX from the menu. Choose the options you want to use and click OK. Entrez Gene ID Field has to be mapped to the node attribute containing the Entrez Gene IDs. After a short period of time you should see an info box stating that the search has completed. New edges should now be visible on your network.
Expanding a node. Select the node (or a small set of nodes) you wish to expand and choose Plugins -> CyEVEX -> Expand nodes with EVEX data. Choose the options you want to use and click OK. Entrez Gene ID Field has to be mapped to the node attribute containing the Entrez Gene IDs. After the search is completed, you should see a new set of nodes and edges on your network. CyEVEX plugin does not change the layout of your network and thus all new nodes will be created to the Cytoscape default spot. Good way to start visualizing your network is to select the new nodes and applying Grid Layout on them.
Linking out to EVEX website. Right-clicking on a node or edge will open a menu which should contain an option called "LinkOut to EVEX". If you selected an edge that has been created with CyEVEX plugin, the detail page of the corresponding event will be opened. Selecting a node or an edge that does not contain EVEX event ID will lead to the main search result page.
Multiple edges can be linked out from Plugins -> CyEVEX -> LinkOut all selected edges. This functionality works only with edges created with CyEVEX.
Searching with gene families. EVEX resource provides homology-based gene family generalizations which can be used also with the CyEVEX plugin. The desired resource of families (e.g. Homologene, Ensembl Genomes) can be selected from CyEVEX settings (Plugins -> CyEVEX -> CyEVEX Options). The created edges are marked with the family in use. While expanding a node with gene families, only interacting families with a gene from the same organism are included in the results. The newly created nodes are identified by the Entrez Gene ID of this organism-specific gene.
Edge attributes
- interaction: coarse interaction type, either Regulation, Indirect_regulation or Binding.
- evex.subtype: refined interaction type, e.g. regulation of expression.
- evex.coarse_polarity: coarse polarity of regulation interaction. Possible choices: Negative, Positive or Unspecified (or Neutral for binding events).
- evex.refined_polarity: refined polarity, interpretation of the real polarity in longer regulation chains.
- evex.confidence: classification confidence of the extracted event.
- evex.generalization: the gene family resource used for finding the given edge
- evex.negation: an estimation if the textual sources counterargument the existence of the given interaction. Value 0 -> the interaction occurs, value 1 -> interaction does not occur
- evex.speculation: an estimation whether the interaction is only speculated instead of a fact. 0 -> considered as a fact, 1 -> speculated
Example network
A Cytoscape session file for the examples below can be downloaded here. This file contains 6 networks, that can be used for testing the plugin.Expanding a node. The example network Mec1, contains only one node which can be used for expanding the network. By selecting this node and choosing Plugins -> CyEVEX -> Expand nodes with EVEX data, 68 interaction partners are found (Figure 1). This network is named as Mec1 expanded in the example .cys file.
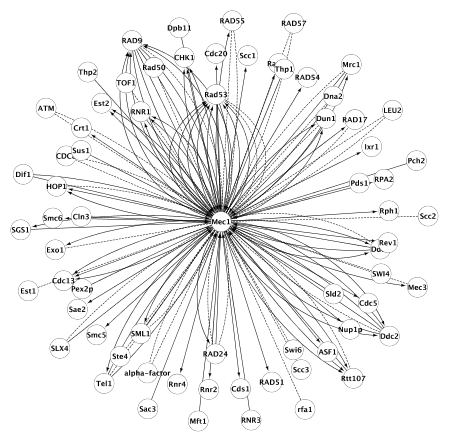
Figure 1: Network created by expanding the one central node (organic layout).
The expansion function can be used iteratively. By expanding some of the previously found nodes, larger network is generated (Figure 2).
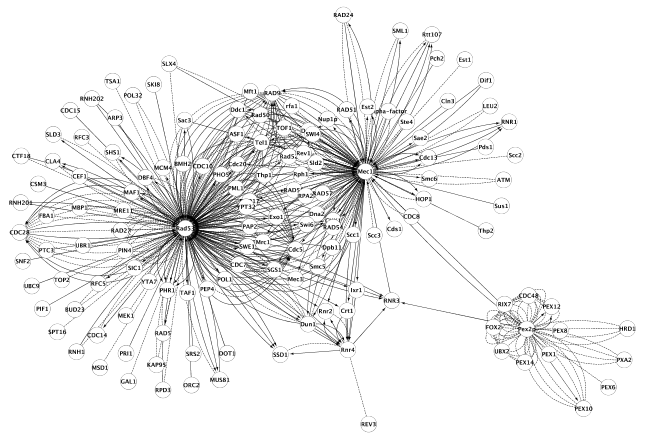
Figure 2: Expanding the network further (organic layout).
Searching edges for existing nodes. Pairwise interactions can be searched by selecting the nodes and choosing Plugins -> CyEVEX -> Search edges from EVEX. This functionality is illustrated in Figures 3 and 4.
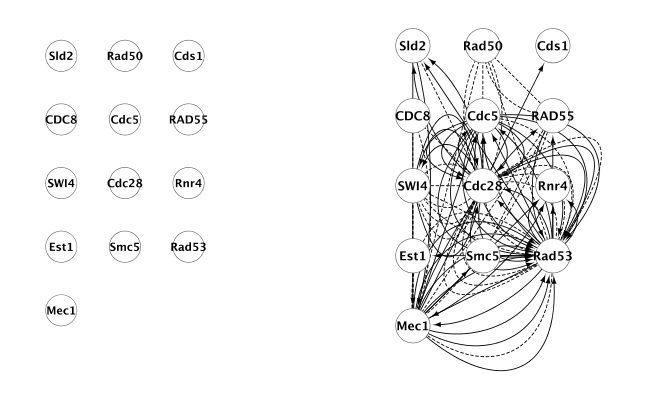
Figure 3: Direct interactions for a set of genes (right). Original node set, without edges, is shown on the left.
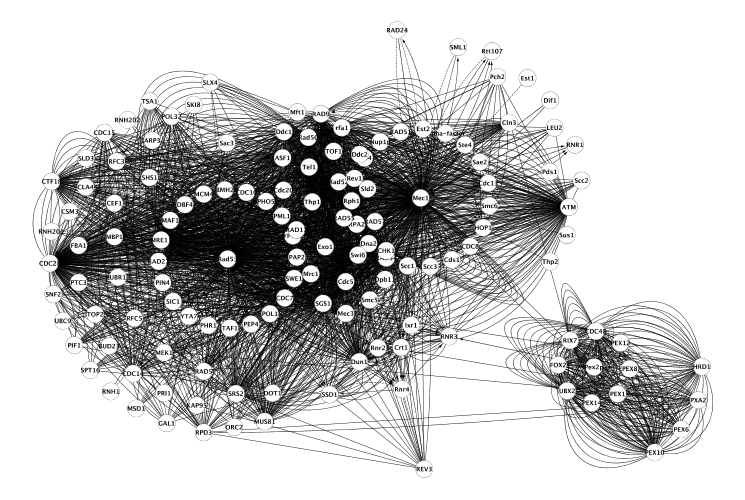
Figure 4: All pairwise interactions for a previously expanded network.
